Frequently Asked Questions
DNA sequencing provides such detailed information that a positive result is generally considered conclusive. If the report indicates that a source of DNA was present in your sample, we can be confident enough in this conclusion there’s little point in estimating an exact probability.
Of course false positives may occur in principle with any test. In this context, possible causes include:
- Database incompleteness. If a novel organism from your sample is not present in the database, but a close relative is present, our process may falsely label the DNA as coming from the relative instead of the unknown organism itself. This is generally an issue for unknown species, and not a problem for well-studied species like known pathogens or parasites.
- Physical contamination. Although we and our clients work hard to prevent contamination while collecting and processing the sample, it seems inevitable that errors must occur at some low frequency in both stages. One benefit of our perspective as we review all the clients’ reports from each batch is that cross-contamination should be apparent. We find no evidence of this. In fact, many times when we report the presence of a parasite or pathogen it’s the only sample in that batch of samples that even contained that subject, so cross-contamination wouldn’t even be possible. With that said, biological contamination is probably the most likely explanation for any false positives that occur.
Its useful to limit our interpretation to what the positive result really says. Its a statement about the DNA present in the sample shipped to us. It may contain DNA from sources that are not alive in your tank; for example, we often find DNA from common food sources like mysis and brine shrimp. It may contain DNA from sources outside the tank, that were not introduced deliberately, including DNA from land plants (presumably introduced via pollen). But the identity of the source of each DNA sequence shown in the report is generally pretty well known, however it got there.
In theory this question generally depends on two factors:
- Sequencing coverage — the number of DNA sequences analyzed from the sample.
- The abundance of the target — at any coverage level, the more abundant targets are more likely to be detected than the rare targets.
We aim to sequence about 10,000 DNA sequences from each sample. In theory, based on binomial probability, we can be 99% confident that we would detect a target that makes up at least 0.05% of the sample. So only the rarest members of the community should escape our test.
In practice, one factor that can complicate this is the distribution of DNA sequences in the sample. If for some reason the sample is swamped with DNA from a single source, this can obscure the presence of rarer targets in the sample. This would be apparent in the sample composition plots in the report, and may call for re-sampling or re-sequencing if it occurs, to ensure we get a high-sensitivity test of your tank.
This is a question that comes up sometimes when clients are hoping to be certain the tank is parasite- or pathogen-free.
On one level the answer is straightforward – we found no evidence of any DNA matching the organism of interest (pathogen, etc). Assuming that we got enough high-quality data from your sample, we expect that if the known parasites or pathogens were present at typical levels, we would have detected them.
(If for some reason we didn’t get enough data from your sample to be confident about this conclusion, we’ll email to discuss – and probably re-run your sample to be sure).
It’s important to recognize that all tests have a limit of detection, so parasites present at a low enough level might be missed. This is true for any testing method. With DNA sequencing, the limit of detection depends on the number of sequences analyzed. For our tests, we analyze thousands of sequences from every client’s sample to maximize the sensitivity of our tests for detecting rare members of the community.
Like any diagnostic test we can’t prove the target is absent, only that we found no evidence of its presence, and that our test would have detected it if it were present at meaningful levels.
What use is a preliminary report?
In some situations, the DNA sequences we get back from a sample are not enough to get a complete picture of the community. These results can still be useful, so we generally put them into a report and label it “preliminary”.
If your report is labeled preliminary: the DNA evidence demonstrates that your sample contains the organisms listed in that report. You can rely on these results just like any DNA sequencing test.
On the other hand, we’re not as confident in these cases about the organisms that we didn’t find in your sample (negative results, e.g. the absence of a parasite). If the report is labeled preliminary, it means there were technical issues with the data that may make the test less sensitive than usual.
Why re-test the sample?
In these cases we sometimes re-run the sample to get a more complete picture of the community in the tank. This isn’t because the preliminary report was wrong. In fact the preliminary report often looks very similar to the result we get back after re-running it.
The reason for re-running these samples is just to double check for the presence of any parasites we might have missed the first time. We want all our clients to get results they can be confident in, so if the data are questionable we sometimes re-run the sample to be sure.
Why upload the preliminary report if it’s not accurate?
The preliminary report is not wrong or inaccurate. Its just likely to miss rare members of the community, which might include parasites or other pests if they are present in the tank.
The preliminary report is just incomplete, but an incomplete picture can still be useful. Sometimes these reports already reveal useful details such as the presence of a parasite, or unusually high levels of dinoflagellates.
We upload these reports because we recognize that our clients’ data are time-sensitive, and we want to deliver their data as soon as they’re available. Its up to clients to decide whether to act on the preliminary information or wait for a re-test of the tank.
In some conditions, Cyanobacteria can form thick mats on rocks and sand in the aquarium. The same process happens periodically in natural coral reefs, but in the aquarium it becomes unsightly and if left untreated can smother and kill corals.
If your sample contains evidence of mat-associated Cyanobacteria, or if you see visible cyanobacterial mats in the aquarium, you might wish to consider the following tips for reducing their abundance.
- Manual removal – we consider this an essential part of any Cyano cleanup strategy. Remove heavily impacted rocks from the tank and scrub thoroughly in a bucket of saltwater. Vacuum clean the substrate using a siphon.
- Increase nitrate levels by about 5-10 ppm. This can be accomplished by directly dosing Sodium Nitrate or Potassium Nitrate, or by increasing the amount or frequency of your feedings with a low-phosphate food.
- Increase microbial diversity by inoculating your tank with a natural product like live rock, live mud or live sand.
There is a surprising amount of variation in the levels of Nitrifying Microbes in saltwater aquariums, beyond what most reef keepers previously imagined. So if your tank shows low levels in the Nitrifying Community section of the report, you’re not alone.
A few things to keep in mind:
- Low levels of nitrifying microbes should only be considered a problem if your aquarium suffers from excess nutrients. This may show up as high levels on your tests, or as nuisance algae.
- So if your nutrient levels are maintained in your desired range using a macroalgal refugium or an algal scrubber, for example, then low nitrifying communities would not be a problem.
- Nitrite-oxidizing bacteria (NOB) are typically much less abundant than ammonia-oxidizing microbes (AOM), so if AOM are low, then NOB are likely to fall below the limits of detection. This should be interpreted as a reduction in the overall nitrifying community rather than a specific deficiency in NOB.
With that said, here are some strategies for enhancing your nitrifying community.
- Most of the popular bottled bacterial products contain one or more types of nitrifying bacteria. So there are many options on the market to supplement these deficiencies, and we don’t endorse any of these products over the others because we have not yet compared their performance ourselves.
- However, the most abundant ammonia-oxidizer in almost all aquariums are not Bacteria, but rather Archaea in the family Cenarchaeaceae. Most bottled products do not contain this group, although Dr Tim’s One & Only does.
- In our experiments, we’ve found that natural materials including Live Rock and Live Mud & Sand are effective for seeding aquariums with a diverse nitrifying community. We expect that materials of this kind from other sources are likely to offer similar benefits.
- Another important option to consider: feed more! The nitrifying community grows or shrinks depending on the amount of available ammonia and nitrite in the water. So if your nitrifying community is low, and you don’t have excess nutrients or nuisance algal growth, increasing either the quantity or frequency of feeding is one of the easiest ways to promote the growth of these populations.
Established, mature reef tanks typically have highly diverse microbial communities, while newer tanks, especially those started using dry rock, typically show much lower scores. Healthy reefs in nature show higher microbial diversity than degraded reefs. These comparisons support the conclusion that high microbial diversity is a positive feature, motivating many reefers to look for ways to increase diversity in their tanks.
In choosing a strategy, it’s worth considering the context to identify the likely reason for this score.
In new tanks, a low score probably indicates that typical community has not been established yet. Inoculation with live rock, live sand or live mud is a good solution in these cases.
In established tanks, a low score may result from the long-term use of sterilizing approaches (UV or Ozone). Unless required for managing known diseases, users may consider limiting their use temporarily so that free-living microbial populations can recover.
In any tanks where the microbial community is dominated by a single type (something microbiologists call a “bloom”), a low diversity score may result from the bloom. In these cases its better to focus on addressing the cause of the bloom (these are often associated with excess nutrients or disease outbreaks) rather than the diversity itself.
Many users ask whether bottled bacterial products are useful for increasing diversity. The difference between a low-diversity tank and a high-diversity tank is measured in the hundreds of different types, while bottled products contain only a few types. Based on these measurements and the long-standing observation that 99% of marine Bacteria cannot be cultured, we consider natural products to be a more effective approach.
A low balance score indicates an atypical microbial community. If this is found in a tank with otherwise unexplained problems, increasing the balance (or adjusting your tank’s microbial community to make it more typical) is recommended.
The strategies for increasing your balance score depend entirely on the specific differences between your tank’s microbiome and the typical tank. The Community Composition section of your report highlights these deviations.
Because these deviations differ from one tank to another, the strategies for increasing your balance score also differ. Adjusting your balance score is accomplished by adjusting the composition of your tank’s microbiome — promoting the growth of some types, inhibiting the growth of others. Your report will include some suggestions customized for your tank.
As you think about strategies for adjusting your tank’s microbiome, it may be useful to start with this overview of the major families present in saltwater aquariums. Your report will also include links to specific information about these groups.
Briefly, you filter water from your aquarium using a syringe-filter, and the biofilm using a sterile swab, using materials included in the sampling kit . Following the instructions in the kit, you then fix the sample, register it into your account and ship it back to us for testing.
See this page for detailed instructions.
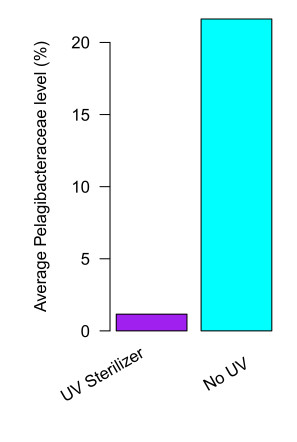
UV sterilizers almost completely eliminate the Pelagibacteraceae
This group is 18 times less abundant on average in tanks with UV than in tanks without. In fact, nearly all tanks with UV sterilizers (98%) have very low Pelagibacteraceae levels (<5%). This effect is so strong we can typically recognize tanks with UV just by looking at their community composition.
Pelagibacteraceae are the most abundant group on natural reefs
A recent study found that Pelagibacteraceae were among the top 3 most abundant families in seawater from coral-dominated communities on the Great Barrier Reef (source). Interestingly, this family was found at lower levels in high-nutrient, inshore habitats dominated by macroalgae. Even in those habitats, however, Pelagibacteraceae remain among of the top 3 most abundant families.
This comparison emphasizes that growing corals in a microbial community without Pelagibacteraceae represents a major deviation from natural conditions.
Recommendations for UV sterilizers in reef tanks
Many reef keepers have found UV sterilizers to be useful tools for controlling outbreaks of nuisance algae, turning the UV on as needed. This seems like a safer and more reasonable use of UV, since the effects of the UV can be monitored visually and it can be turned off again once the problem is resolved.
Some aquarists run UV preventatively in an effort to prevent disease outbreaks. We advise against this practice for reef tanks, since it alters the microbial community so dramatically.
Overall we view UV as similar to a strong dose of antibiotics – not something to be administered constantly, but used as needed for especially difficult infections (or nuisance algae).
The high-throughput DNA sequencing technologies that make this analysis possible require that we process samples in large batches (50-200 at a time). Because a sample received early in a batch may have to wait a while until the batch is complete, this can introduce delays.
You can generally expect to receive a detailed analysis of your sample within 4 weeks of its arrival at our facility. Depending on your position in the queue, you may receive your report a bit earlier or later than this.
If you’re curious about what’s happening with your sample while you wait, here is a brief explanation of what happens:
The way this technology works requires that we process samples in batches. So when we receive your sample, we extract the DNA, and prepare it for DNA sequencing. This takes about a week of hands-on time in the lab to process each batch.
Once we accumulate enough samples for sequencing, we send the batch to the DNA sequencing facility. DNA sequencing is typically completed within 1 week.
When sequencing is completed, they send us the raw data (~2 million DNA sequences). We run our programs to analyze the data, and within about 12 – 24 hours your report is ready. At that time, you’ll get an email with a link to your results.
So overall the turnaround time for these samples depends on when your sample arrives at our facility. If it arrives right before we begin processing a batch, it should be completed in less than 2 weeks. Sample arriving after that will be processed in the following batch, generally starting about 2 weeks later.
Because this process includes several variables outside of our control, we can’t make any guarantees about timing. But we hope this description helps you understand the reasons for the time it takes to process your samples.
If you have questions at any point during the process feel free to contact us by email.
Our rates and payment schedules are shown at https://aquabiomics.com/place-an-order.
Depending on the number of samples you test at once, the test ranges from $99 (for a single sample) to $49 per sample (for 10 or more samples). These discounts are “built in”, so you can add multiple tests and see the adjusted price in your shopping cart.
You also have the option to pay for multiple tests at once, or schedule recurring tests for a discounted rate.
Here are some commonly asked questions about the reports.
- What does a negative result mean?
- How sensitive is the test?
- Can the test give false positives?
- What does it mean if an organism is listed multiple times in the report?
- Why does my report contain DNA from land plants or animals?
This page is a work in progress. Please contact us by email if you have any suggestions for questions we should add to the list!
- Register an account and log in at https://aquabiomics.com/login
- Choose the number of tests and payment schedule at https://aquabiomics.com/place-an-order
- If you have a coupon code, you can apply this in the shopping cart to see your cost before ordering.
There is an established process for designating a bacterium as a pathogen that requires completing a series of experimental steps (e.g. infecting healthy corals with the pathogen and documenting the appearance of disease symptoms). Some of these steps are especially challenging with reef-building corals, so the list of “official” coral pathogens can lag behind the state of our knowledge about likely pathogens.
In this recently added section of the microbiome report, we screen for several suspected pathogens associated with known coral diseases. There is evidence linking these bacteria to coral diseases, but they have not been formally designated as pathogens yet.
Brown Jelly Disease (BJD)
This disease primarily affects Euphyllia corals including hammers, frogspawns, and torches. Like most coral diseases the name describes the symptoms – in this case, the polyp and internal tissues dissolve into a brown gelatinous blob. By the time these symptoms are visible, the symptomatic polyps are doomed and nearby polyps or other colonies are often already infected.
We’ve identified an unclassified member of the Arcobacter genus that is associated with BJD in Euphyllia corals. This is reported as “Arcobacter sp. type 1103”. Infections with this bacterium have been successfully treated using low doses of Ciprofloxacin. You can read more about this here.
Stony Coral Tissue Loss Disease (SCTLD)
This disease has recently emerged as a major threat to corals throughout the Caribbean region. Researchers have recently completed an extensive study of the microbiome in corals affected by the disease, and identified a group of bacteria associated with SCTLD. Several of these same bacterial types show up occasionally in reef tanks, and are included in this section of the report.
What does it mean for your tank?
First consider the prevalence and average level of the bacterium. The presence of a suspected pathogen would raise greater concerns if the pathogen is occurs rarely (low prevalence) or present at higher than usual levels. A suspected pathogen that occurs frequently or is present at typical low levels would raise less concern.
Also consider the affected species. Brown Jelly Disease is primarily an issue for Euphyllia corals. SCTLD affects a wide range of LPS corals. Concerns about these suspected pathogens will depend on whether the affected species are present in your tank.
What can be done about it? In principle both diseases are treatable with antibiotics, although the specific treatment protocols remain a work in progress. We’ve described an in-tank treatment for BJD using low doses of Ciprofloxacin. Researchers studying SCTLD have used a topical antibiotic putty to treat diseased corals in the field.
Aquarists considering antibiotic treatments should consider the following:
- Antibiotics should only be used therapeutically, never as a preventative measure. The routine preventative use of antibiotics leads to the emergence of antibiotic resistance in microbial communities, which can lead to serious health risks.
- Antibiotic treatments are likely to impact beneficial microbes in addition to pathogens. This can be minimized by the choice of antibiotics and dosages. Testing the microbiome can help users determine whether supplementation is needed to restore the community after an especially aggressive antibiotic treatment.
Bacterial pathogens are common in fish aquaculture but have been rarely diagnosed in the aquarium hobby. Our tests have revealed that a few known pathogens show up in a small fraction of hobbyist tanks. The most commonly encountered are Vibrio fortis and Photobacterium damselae.
Both of these pathogens seem to affect a limited range of fish in the aquarium hobby (V. fortis is a pathogen of seahorses and their relatives, while P. damselae affects a somewhat broader range of fish including the Damsel family). So the fish in your tank may not be susceptible. Some types of P. damselae are more pathogenic than others, and our test cannot distinguish between these types. So its possible that the type present in your tank could be a less pathogenic variety.
Despite these uncertainties, most reefkeepers would be rightly concerned to find a known fish pathogen in their tank. We’ve started a discussion on the topic here.
While we cannot offer specific advice for treating your fish (which would constitute veterinary advice), our practice is to remove symptomatic animals from the tank whenever possible, transferring them to a separate quarantine tank (QT) for observation. This removes a major reservoir of the pathogen from your tank.
If you choose to treat fish, this can be more easily done in the QT. You can find an excellent discussion on some of the options for treating bacterial infections in saltwater fish here.
We generally do not recommend antibiotic treatments on the display tank, for a variety of reasons. If you find a bacterial pathogen in your tank, we suggest the following general strategy, modified as needed depending on the details of the situation.
- Remove the main source of the pathogen from your tank. This may involve transferring a symptomatic fish to QT or trimming dead parts off a coral colony. Better to lose an individual fish or coral than all of them!
- Deep clean the tank. Many pathogens persist in biofilms and sediments. Scrub the glass, vacuum the substrate, and do a series of relatively large water changes (up to 50% of the tank volume).
- Maintain relatively low nutrients (no more than 5 ppm nitrate) for a while to avoid feeding a bloom of pathogenic Bacteria. Maximize nutrient export (e.g. run your skimmer or refugium lights for a longer period).
If your fish don’t show any symptoms, its perfectly reasonable to do nothing. Some groups of fish are less susceptible to particular pathogens, and individual fish can develop resistance over time. A pathogen in your tank may pose little risk to the fish that currently exist in your tank, but a major risk for susceptible new introductions. In that case, you may wish to avoid adding fish from known susceptible groups while the pathogen remains in your tank.
The Diversity score is simply the number of different types of Bacteria or Archaea in your sample, based on differences in their DNA sequences. This is sometimes called alpha diversity. Diversity does not take into account any differences in the relative abundance of the various types; it simply counts them.
This concept is easy to visualize with large animals like aquarium fish. A tank housing only a single type of fish has low animal diversity, while a reef tank with multiple species of fish, snails, crabs, and corals has high animal diversity.
We can describe microbial communities in the same way, although here the diversity can’t be seen with the naked eye. A diverse microbial community includes many different kinds of microbes (single celled organisms including Bacteria and Archaea). A pure culture of a single bacterial type, on the other hand, has very low diversity.
Technical details: because the raw number of types detected in a sample is affected by the total number of DNA sequences in the sample, we account for variation in DNA sequencing coverage using a statistical approach called rarefaction. This allows for an apples-to-apples comparison between samples that receive different levels of sequencing coverage.
The Balance score is based on comparing the relative abundance of the major microbial families in your sample with those in the typical saltwater aquarium. This score describes whether your sample has a typical community (higher scores) or an unusual community (lower scores).
A high score indicates that the major families of Bacteria present in typical saltwater tanks are present at similar levels in your tank. A low score indicates that one or more of these families is present at very different levels than in the typical tank.
Taken by itself, a low score should not necessarily be interpreted as a reason for alarm. But it provides a conclusive answer to the question, “Is the microbial community in my tank normal or abnormal?” If there are unexplained problems in the aquarium ecosystem, a low score suggests that disruptions in the aquarium microbiome could be playing a role.
Unfortunately, we are unable to offer our testing services outside of the US at this time.
Vibrio fortis
Prevalence
V. fortis is the bacterial fish pathogen most commonly detected in reef tanks.
Vibrio fortis is the most prevalent pathogen in saltwater aquariums, occurring in over half the tanks we’ve sampled, usually without any reported symptoms in fish.
To our knowledge this pathogen has only been associated with disease in seahorses and their relatives [1]. In laboratory challenge experiments, however, it also proved pathogenic for rainbow trout and brine shrimp, suggesting a much wider range of susceptible hosts [2].
- Wang, X., et al. “A Novel Pathogenic Bacteria (Vibrio Fortis) Causing Enteritis in Cultured Seahorses, Hippocampus Erectus Perry, 1810.” Journal of fish diseases 39.6 (2016): 765-769.
- Austin, Brian, et al. “Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii.” Environmental microbiology 7.9 (2005): 1488-1495.
Our Aquarium Microbiome Testing service includes:
- Identification and classification of the different kinds of microbes in your aquarium
- Measurements of each type’s relative abundance in the community
- Comparison with the microbial communities found in typical reef tanks
- Measurements of beneficial microbes, including nitrifying microbes that improve water quality
- Screening for specific bacterial pathogens of fish and corals

