The Core Microbiome of a Saltwater Aquarium
What’s in the microbiome of a saltwater aquarium?
Aquarists have known for a long time that microbes played important roles in our aquariums, but have been unable to directly measure the microbial communities in their tanks until recently.
To better understand these microbial communities, I’ve teamed up with a group of local hobbyists and coral growers to sample microbial communities in a wide variety of tanks. In my new lab at AquaBiomics I used DNA sequencing to identify the different kinds of microbes present in each tank, and measure their relative abundance.
By comparing these communities across different tanks, I identified a core set of families found in nearly all aquariums we sampled. My goal in this report is to introduce these families of microbes, for aquarists interested in learning more about the microbes living in their own tanks.
How DNA sequencing is used to study the microbiome
For this study, my collaborators sampled seawater from multiple tanks using custom sampling kits I provided, then shipped the samples back to my lab at AquaBiomics. Altogether we sampled 20 tanks operated by 7 people, ranging from home display tanks to coral aquaculture facilities.
We chose to sample microbial populations in the water because this can be sampled more consistently across tanks than other strategies (e.g. sand, rock, etc.) I view this as analogous to sampling a patient’s blood to diagnose issues with their heart or liver. Our approach is supported by a recent study which found that water samples are ideal for measuring ecosystem disturbances on natural coral reefs.
Very few (probably <1%) of microbes can be grown in culture, so many researchers now use DNA sequencing to study these communities. I extracted DNA from each sample, and sequenced a genetic marker called 16S that’s been widely used for studies of microbial communities in natural habitats. To ensure that we sampled the community deeply enough to detect rare microbes, I analyzed about 10,000 DNA sequences per sample. By comparing these sequences with public databases, I identified the type of microbe from which each DNA sequence was extracted, and counted these sequences to evaluate the relative abundance of each type.
Established saltwater aquariums support diverse microbial communities
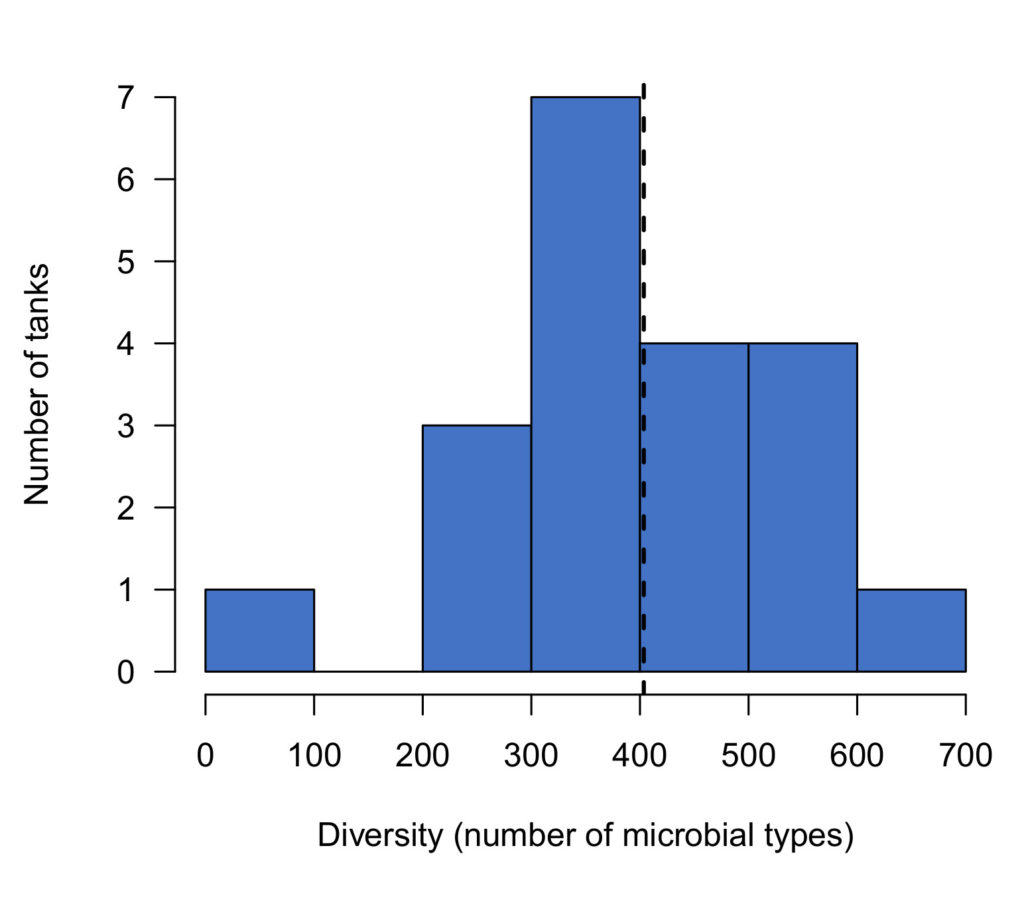
These data revealed that the water circulating through a typical saltwater aquarium contains a complex mixture with hundreds of different kinds of microbes. In our survey of hobbyist and aquaculture systems we found 400 different types per tank on average.
The variation across aquariums was striking; the most diverse tank we sampled contained more than 7 times as many microbial types as the least diverse. Most tanks (80%) contained at least 300 microbial types, but some contained far fewer (Figure 2).
Why does this diversity matter? After all, as aquarists we aren’t really interested in the names of the microbes, as much as we are in what they can do for us.
Classifying each microbe to the family level reveals that the hundreds of different types in each sample represented a large number of different families (90 different families per sample, on average).
These families have have different, characteristic metabolic activities and play different roles in nature and presumably in our aquariums. So this diversity is not just a matter of DNA sequences; the different kinds of microbes do different things.
Aquarists have often debated what makes a tank “mature”. From a microbial perspective, our analysis of established, mature saltwater aquariums establishes a target range for microbial diversity, providing an objective metric for determining the maturity of a new tank.
The aquarium water microbiome is dominated by a few families
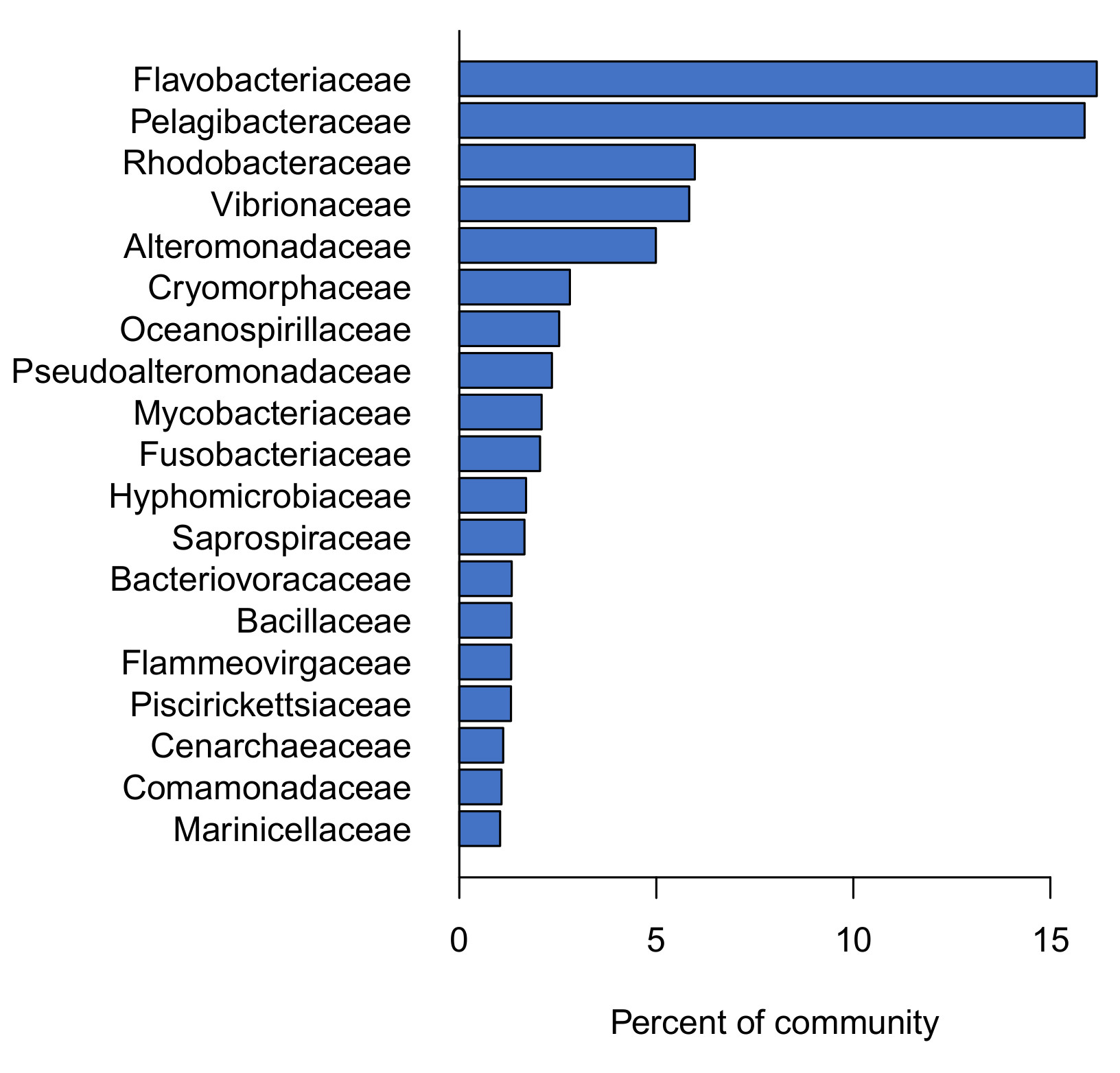
In addition to identifying the kinds of microbes present in each sample, DNA sequencing provides information on each types’ relative abundance (in other words, the ratios between the different types). This analysis revealed that a relatively short list of families accounted for the majority of the aquarium water microbiome (Figure 3).
These observations demonstrate that there is a core microbiome characteristic of mature saltwater aquariums. These families were present in almost every tank we sampled, and made up a relatively large fraction of the community in each tank. Disruptions of this core microbiome may contribute to otherwise unexplained problems like reduced water quality, nuisance algae, and coral or fish diseases.
To identify the core saltwater aquarium microbiome, we focused on the set of 19 families that each made up at least 1% of the community, on average. Together, these “core” families accounted for 73% of the total community. Two of these were especially abundant (Pelagibacteraceae and Flavobacteriaceae), each accounting for about 16% of the total community (Fig 3). The other “non-core” families (158) were each present in fewer tanks and at very low levels (0.17% of the community on average).
In the remainder of this article, we focus on describing these core families that account for the majority of the water microbiome across almost all tanks we sampled.
The 19 microbial families in the core aquarium microbiome
Each of these core families is described in the following table. These descriptions focus on marine members of each family, when specific information was available about marine groups. (Note: if this table does not display well on your mobile device, consider switching to the Desktop version of the site to view this table.)
Table 1
Microbial families of the core saltwater aquarium microbiome.| Family | Description | Metabolic capabilities | Ecological roles & responses | |
|---|---|---|---|---|
| 1 | Flavobacteriaceae | Gram-negative, rod-shaped, non-motile or gliding Bacteria (Bacteroidetes) | Generally aerobic & chemoheterotrophic | Most diverse family in Bacteroidetes; occurs in essentially all habitats. Specialized for degrading polysaccharides & proteins. Often the most abundant group in aquatic habitats. Frequently associated with surfaces, including animals, macroalgae, or detritus. |
| 2 | Pelagibacteraceae | Gram-negative, rod-shaped, free-living Bacteria (Alphaproteobacteria) | Aerobic & chemoheterotrophic | Previously called SAR11, this is thought to be the most abundant bacterial group in the ocean worldwide. Well-adapted for life in the low-nutrient waters of the open ocean. Require reduced sulfur compounds, glycine, and dissolved organic carbon for growth. |
| 3 | Rhodobacteraceae | Gram-negative Bacteria (Alphaproteobacteria), mostly rod-shaped, some free-living | Mostly aerobic & chemoheterotrophic, some photoheterotrophic | Extremely diverse, widely distributed and highly abundant in marine habitats including open ocean, sediments, and algal biofilms. Degrade sulfur-containing compounds (e.g. sulfite, DMSP). Many use methylated amines (MA) as primary nitrogen source. |
| 4 | Vibrionaceae | Gram negative, motile, Bacteria (Gammaproteobacteria); curved or straight rod-shaped | Aerobic or anerobic; chemoheterotrophic, photoautotrophic, or chemoautotrophic; some biolumenescent. | Widely distributed in marine habitats, including many associations with animals. This family includes many human or animal pathogens, including bacteria that can cause wound infection from exposure to contaminated water. |
| 5 | Alteromonadaceae | Gram-negative, rod-shaped, motile Bacteria (Gammaproteobacteria) | Aerobic & chemoheterotrophic | Widely observed in seawater samples. Can use a broad range of dissolved nutrients including sugars and amino acids, and blooms in high glucose conditions. |
| 6 | Cryomorphaceae | Gram-negative, rod-shaped or filamentous Bacteria (Bacteroidetes). Non-motile or gliding. | Aerobic or facultatively anerobic; chemoheterotrophic. | Primarily marine microbes, with some freshwater members. Generally surface-associated. Not primary degraders, but contribute to secondary production. Metabolizes amino acids and other organic acids. Nutritonal requirements remain poorly defined, but supported by organic extracts (e.g. yeast). |
| 7 | Oceanospirillaceae | Gram-negative, spiral- or rod-shaped, motile Bacteria (Gammaproteobacteria) | Aerobic & chemoheterotrophic | Almost exclusively marine. Grows on amino acids, other organic acids, and ammonia. Contributes to biofilm communities, and growth is stimulated by nutrient enrichment (C, N, & P). |
| 8 | Pseudoalteromonadaceae | Gram-negative, rod-shaped or round, motile Bacteria (Gammaproteobacteria) | Aerobic & chemoheterotrophic | Ecologically important in a wide variety of marine habitats. Produce a variety of bioactive compounds, including many antimicrobial or antiviral comounds. Plays important roles in the formatin of biofilms. Can inhibit establishment and growth of algae. High molecular weight DOM promotes growth of this family. |
| 9 | Mycobacteriaceae | Not truly Gram-positive or negative, rod-shaped, non-motile Bacteria (Actinobacteria) | Aerobic; mostly chemoheterorophic | Grows on a variety of simple sugars, alcohols, or hydrocarbons. Growth is promoted by addition of fatty acids. Not generally pathogenic or symbiotic, but includes a few very important human pathogens (leprosy, tuberculosis). Includes the aquarium-related pathogen M. marinum (‘fish-tank granuloma’). |
| 10 | Fusobacteriaceae | Gram-negative, rod-shaped or round, non-motile Bacteria (Fusobacteria) | Anaerobic or microaerophilic, chemoheterotrophic | Occurs in a variety of habitats. Ferments organic nutrients including carbohydrates, amino acids, and peptides. Found in sediments and associated with animals. |
| 11 | Hyphomicrobiaceae | Gram-negative Bacteria (Alphaproteobacteria) with round to rod-shaped cells, some motile | Includes chemoheterotrophic, methylotrophic, chemolitoautotrophic, and photosynthetic | Found in essentially every habitat. Grows on organic acids and sugars. |
| 12 | Saprospiraceae | Gram-negative rod-shaped Bacteria (Bacteroidetes), some show gliding motility | Aerobic & chemoheterotrophic | Primarily marine, some freshwater. Typically associated with sediments, multicellular organisms, or other surfaces. Capable of breaking down and living on complex macromolecules (e.g. polysaccharides, proteins). Some prey on other bacteria or algae, suggesting a role for this group in controlling algal growth on surfaces. |
| 13 | Bacteriovoracaceae | Gram-negative, rod-shaped, motile Bacteria (Deltaproteobacteria) | Aerobic or anerobic; chemoheterotrophic | Widely distributed across aquatic and terrestrial ecosystems. Obligate predators of other gram-negative bacteria. Play important roles in controlling microbial community size and diversity. |
| 14 | Bacillaceae | Gram-positive, rod-shaped, motile Bacteria (Firmicutes) | Aerobic or anaerobic; chemoheterotrophic | The hardiest and mos widely distributed group of bacteria. Spore-forming. Found throughout aquatic and terrestrial habitats, often in association with plants or animals. Primarily saprophytic. Plays important roles in nutrient cycling. Capable of degrading and living on complex macromolecules or simple sugars. Blooms rapidly in response to nutrient addition. |
| 15 | Flammeovirgaceae | Gram-negative, rod-shaped Bacteria (Bacteroidetes) | Aerobic or anaerobic; chemoheterotrophic | Occurs in both terrestrial and aquatic habitats. Commonly observed in marine sediments. Litle information is available on their activity. |
| 16 | Piscirickettsiaceae | Gram-negative, rod-shaped, Bacteria (Gammaproteobacteria), some motile | Aerobic & chemoheterotrophic | A diverse group with a broad range of activities. Includes methylotrophic bacteria with important roles in carbon cycles, and a fish pathogen Piscirickettsia salmonis |
| 17 | Cenarchaeaceae | Round or rod-shaped Archaea (Thaumarchaeota), some motile | Aerobic, chemoautotrophic | Found in essentially all habitats including extreme environments. Important ammonia-oxidizing activities, especially when ammonia levels are low; ammonia-oxidizing Archaea consume more ammonia than AOB. |
| 18 | Comamonadaceae | Gram-negative, round or rod-shaped Bacteria (Betaproteobacteria), some motile | Generally aerobic heterotrophic; many exceptions | A large and diverse group that includes a wide range of lifestyles. Occurs in soil and water samples from a wide range of habitats, and in association with plants or animals. Most are free-living saprophytes. Some grow autotrophically on hydrogen or nitrate. |
| 19 | Marinicellaceae | Gram-negative, rod-shaped, non-motile Bacteria (Gammaproteobacteria) | Aerobic & chemoheterotrophic | Newly described family occuring in seawater samples. Little information is available on its ecological roles. Requires salt and organic nutrients (e.g. hydrolyzed proteins) for growth. |
Microbes of special interest for aquarists
Table 2
The average abundance of microbial groups of interest in a typical saltwater aquarium.| Group | Types found | Average % of community |
|---|---|---|
| Ammonia-oxidizing Bacteria | Nitrosomonadaceae, Nitrosococcus | 0.67% |
| Ammonia-oxidizing Archaea | Cenarchaeaceae | 0.85% |
| Nitrite-oxidizing Bacteria | Nitrobacter, Nitrospinaceae, Nitrospiraceae | 0.14% |
| Cyanobacteria | Acaryochloridaceae, Phormidiaceae, Pseudanabaenaceae, Synechococcaceae, Ulvophyceae, Xenococcaceae | 0.32% |
| Fish pathogens | Photobacterium damselae, Piscirickettsia salmonis | 0.07% |
| Coral pathogens | none | 0.00% |
Summary
- The water in a typical reef tank contains hundreds of different types of microbes with diverse metabolic capabilities.
- The aquarium microbiome is dominated by a core set of 19 families that are relatively abundant in most tanks.
- Bacteria made up most of the aquarium microbiome, but the small number of Archaea include types with important roles in nutrient processing
- Sampling the water of a saltwater aquarium allows for detection of specific beneficial microbes or pathogens
- The differences in metabolic capabilities and nutritional requirements in the core saltwater aquarium microbiome suggest that the microbiome composition can affect dissolved nutrient levels, and vice versa.

